8.1. Pharmacophore 3D Visualization
The script gen_ph4_3d_vis.py generates 3D visual representations of pharmacophores read from an input file and writes the resulting vector graphics data to one or more (in case of multiple input pharmacophores) output file(s).
Synopsis
python gen_ph4_3d_vis.py [-h] -i <file> -o <file> [-c] [-d] [-t]
Mandatory options
- -i <file>
Input pharmacophore file (.pml, .cdf)
- -o <file>
3D visualization data output file (.r3d, .stl, .ply, .wrl)
Other options
- -h, --help
Show help message and exit
- -c
Do not output spheres representing the feature centers (default: false)
- -d
Make vector and plane features undirected (default: false)
- -t
Disable feature transparency (default: false)
Example
Generation of a Raster3D file for the pharmacophore of ligand MIT (complex 1DWC ):
$ python gen_ph4_3d_vis.py -i 1dwc_MIT_ph4.pml -o 1dwc_MIT_ph4.r3d
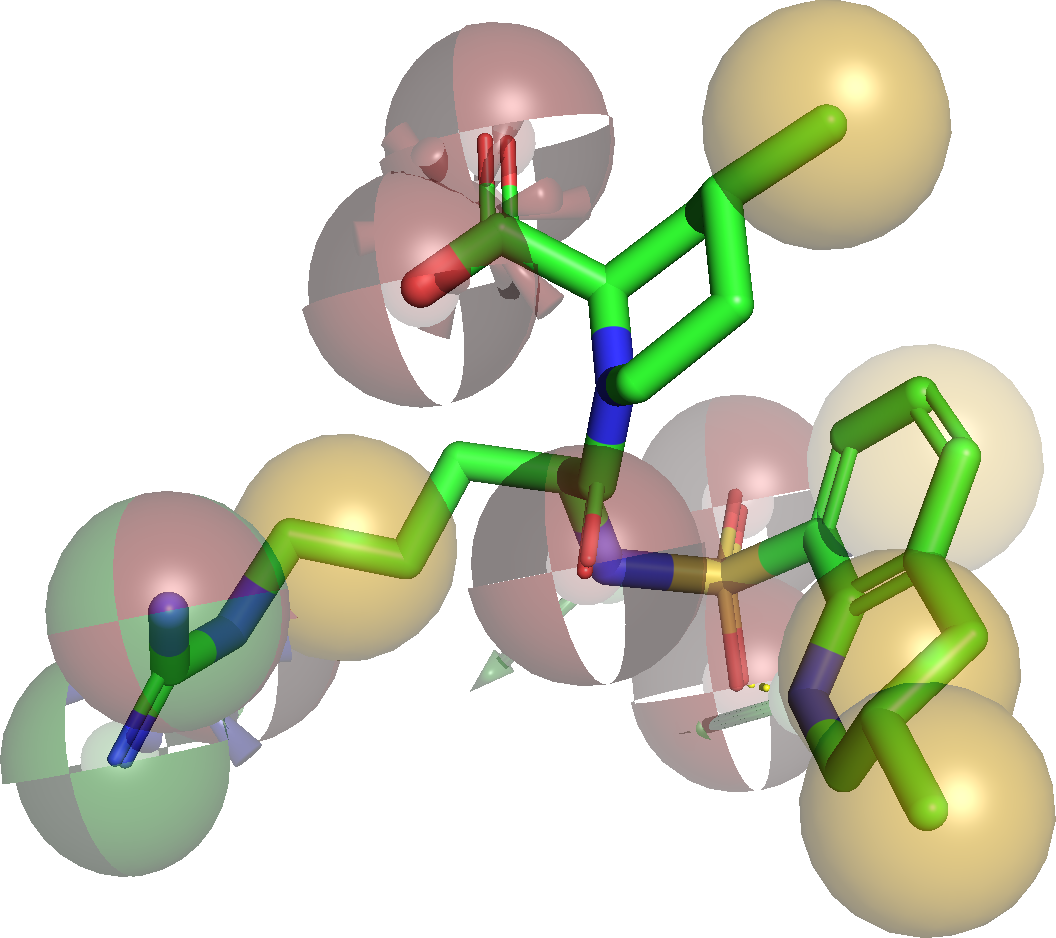
PyMOL displaying the structure and pharmacophore of ligand MIT
Code
1import sys
2import os
3import argparse
4
5import CDPL.Util as Util
6import CDPL.Pharm as Pharm
7import CDPL.Vis as Vis
8
9
10def parseArgs() -> argparse.Namespace:
11 parser = argparse.ArgumentParser(description='Generates 3D vector graphics representations of pharmacophores for the display by external programs.')
12
13 parser.add_argument('-i',
14 dest='in_file',
15 required=True,
16 metavar='<file>',
17 help='Input pharmacophore file (*.pml, *.cdf)')
18 parser.add_argument('-o',
19 dest='out_file',
20 required=True,
21 metavar='<file>',
22 help='3D visualization data output file (*.r3d, *.stl, *.ply, *.wrl)')
23 parser.add_argument('-c',
24 dest='no_ftr_ctrs',
25 required=False,
26 action='store_true',
27 default=False,
28 help='Do not output spheres representing the feature centers (default: false)')
29 parser.add_argument('-d',
30 dest='no_ftr_dir',
31 required=False,
32 action='store_true',
33 default=False,
34 help='Make vector and plane features undirected (default: false)')
35 parser.add_argument('-t',
36 dest='no_transp',
37 required=False,
38 action='store_true',
39 default=False,
40 help='Disable feature transparency (default: false)')
41
42 return parser.parse_args()
43
44def main() -> None:
45 args = parseArgs()
46
47 # create reader for input pharmacophores (format specified by file extension)
48 reader = Pharm.PharmacophoreReader(args.in_file)
49
50 # create an instance of the default implementation of the Pharm.Pharmacophore interface
51 ph4 = Pharm.BasicPharmacophore()
52
53 # instantiate class implementing the creation of visual 3D
54 # representations of Pharm.FeatureContainer objects
55 ph4_3d_repr_factory = Vis.FeatureContainerObject3DFactory()
56
57 # apply the value of option -c
58 Vis.setShowFeatureCentersParameter(ph4_3d_repr_factory, not args.no_ftr_ctrs)
59
60 # process option -t
61 if args.no_transp:
62 ftr_colors = Vis.DefaultFeatureColorTable()
63
64 for color in ftr_colors.values():
65 color.setAlpha(1.0)
66
67 Vis.setFeatureColorTableParameter(ph4_3d_repr_factory, ftr_colors)
68
69 out_file, out_file_ext = os.path.splitext(args.out_file)
70
71 # read and process pharmacophores one after the other until the end of input has been reached
72 try:
73 i = 1
74
75 while reader.read(ph4):
76 try:
77 if args.no_ftr_dir: # if pharm. features shall be output without orientation information (option -d)
78 Pharm.clearOrientations(ph4) # clear orientation property of all features
79 Pharm.removePositionalDuplicates(ph4) # and reduce multiple features having the same type and position to one feature
80
81 # create writer for the output file (file format determined by the file extension)
82 writer = Vis.Object3DWriter(f'{out_file}_{i}{out_file_ext}')
83
84 # generate the 3D visual representation of the read pharmacophore
85 ph4_3d_repr = ph4_3d_repr_factory.create(ph4)
86
87 # output the generated visualization data
88 if not writer.write(ph4_3d_repr):
89 sys.exit('Error: output of pharmacophore visualization data failed')
90
91 i += 1
92 writer.close()
93
94 except Exception as e:
95 sys.exit('Error: generation or output of pharmacophore visualization data failed:\n' + str(e))
96
97 except Exception as e: # handle exception raised in case of severe read errors
98 sys.exit('Error: reading pharmacophore failed:\n' + str(e))
99
100 sys.exit(0)
101
102if __name__ == '__main__':
103 main()